Peptidoglicano: Diferenzas entre revisións
Sen resumo de edición |
|||
| Liña 10: | Liña 10: | ||
== Estrutura == |
== Estrutura == |
||
A capa de peptidoglicano na parede celular bacteriana é unha estrutura que forma unha [[rede cristalina]] constituída por cadeas liñais paralelas de dous [[aminoazucree]]s alternantes, a [[N-acetilglicosamina|''N''-acetilglicosamina]] (GlcNAc ou NAG) e o [[ácido N-acetilmurámico|''N''-acetilmurámico]] (MurNAc ou NAM), os cales están unidos por [[enlace glicosídico]] β-(1,4). Cada residuo de MurNAc está unido a un curto [[péptido]] de 4 a 5 residuos de aminoácidos, que contén [[alanina|<small>L</small>-alanina]], [[ácido glutámico|ácido <small>D</small>-glutámico]], [[ácido meso-diaminopimélico|ácido ''meso''-diaminopimélico]], e [[alanina|<small>D</small>-alanina]] no caso de ''[[Escherichia coli]]'' (unha bacteria Gram-negativa) ou [[alanina|<small>L</small>-alanina]], [[glutamina|<small>D</small>-glutamina]], [[lisina|<small>L</small>-lisina]], e <small>D</small>-alanina cunha ponte peptídica de 5 [[glicina]]s que une os tetrapéptidos no caso de ''[[Staphylococcus aureus]]'' (unha bacteria Gram-positiva). Estes aminoácidos, excepto os <small>L</small>-aminoácidos, non aparecen en proteínas, polo que poderían ser unha protección contra o ataque da maioría das [[peptidase]]s. |
|||
Os enlaces cruzados entre os aminoácidos de diferentes cadeas de aminoazucres establécense pola acción do encima [[transpeptidase]] e orixinan unha estrutura tridimensional forte e ríxida. A secuencia específica de aminoácidos e a estrutura molecular varía coa especie bacteriana considerada. <ref>{{cita libro | autor = Ryan KJ, Ray CG (editors) | título = Sherris Medical Microbiology | edición = 4th | editor = McGraw Hill | ano = 2004 | isbn = 0-8385-8529-9 }}</ref> |
|||
<gallery widths="340" heights="240"> |
<gallery widths="340" heights="240"> |
||
Ficheiro:Mureine.svg|Estrutura do peptidoglicano. |
|||
Ficheiro:Gram-positive cellwall-schematic.png|Parede celular Gram-positiva. |
|||
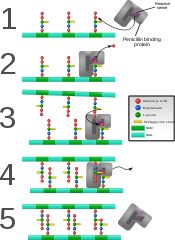
Ficheiro:PBP catalysis.svg|[[Proteína de unión á penicilina]] formando enlaces cruzados nunha parede bacteriana acabada de formar. |
|||
</gallery> |
</gallery> |
||
Revisión como estaba o 1 de xuño de 2012 ás 17:34
Este artigo está a ser traducido ao galego por un usuario desta Wikipedia; por favor, non o edite. O usuario Miguelferig (conversa · contribucións) realizou a última edición na páxina hai 11 anos. Se o usuario non publica a tradución nun prazo de trinta días, procederase ó seu borrado rápido. |

O peptidoglicano, tamén chamado mureína, é un polímero formado por azucres e aminoácidos que forma unha capa que rodea a membrana plasmática da maioría das bacterias (pero non das arqueas) e que constitúe a súa parede celular. O compoñente carbohidrato do peptidoglicano é un heteropolisacárido formado por residuos alternantes dos monosacáridos N-acetilglicosamina e ácido N-acetilmurámico, unidos entre si por enlace glicosídico β-(1,4). As cadeas deste heteropolisacárido dispóñense paralelamente. Unido ao ácido N-acetilmurámico está o compoñente aminoacídico, que é un péptido de tres a cinco aminoácidos. Esta cadea peptídica pode establecer enlaces cruzados cos péptidos doutras cadeas do heteropolisacárido, formando unha capa reticular tridimensional. [1]
Algunhas Archaea teñen unha capa bastante similar pero de pseudopeptidoglicano ou pseudomureína, na que os residuos de azucres están unidos por enlaces glicosídicos β-(1,3) e os azucres que a forman son a N-acetilglicosamina e a ácido N-acetiltalosaminurónico. Isto explica que a parede das arqueas sexa insensible á lisocima, que ataca ao peptidoglicano. [2]
O peptidoglicano exerce un papel estrutural na célula bacteriana, dándolle forza estrutural, e protexéndoa da presión osmótica exercida polo citoplasma. Aínda que o peptidoglicanolle dá forza estrutural á parede, para a determinación da súa forma necesítanse tamén as proteínas MreB e RodZ. [3][4] O peptidoglicano está tamén implicado na fisión binaria durante a división celular bacteriana.
A capa de peptidoglicano é moito máis grosa nas bacterias Gram-positivas, nas que ten entre 20 e 80 nanómetros de grosor, ca nas Gram-negativas, nas que só ten entre 7 e 8 nanómetros, coa adhesión dunha capa S. O peptidoglicano constitúe arredor do 90 % do peso seco das bacterias Gram-positivas, pero só o 10% das Gram-negativas. Deste modo, a presenza de grandes cantidades de peptidoglicano é a característica principal para caracterizar as bacterias Gram-positivas. [5] Nas bacterias Gram-positivas o peptidoglicano é importante para a adhesión a superficies e para o estereotipado. [6] As partículas de aproximadamente 2 nanómetros poden atravesar o peptidoglicano, tanto nas Gram-positivas coma nas negativas. [7]
Estrutura
A capa de peptidoglicano na parede celular bacteriana é unha estrutura que forma unha rede cristalina constituída por cadeas liñais paralelas de dous aminoazucrees alternantes, a N-acetilglicosamina (GlcNAc ou NAG) e o N-acetilmurámico (MurNAc ou NAM), os cales están unidos por enlace glicosídico β-(1,4). Cada residuo de MurNAc está unido a un curto péptido de 4 a 5 residuos de aminoácidos, que contén L-alanina, ácido D-glutámico, ácido meso-diaminopimélico, e D-alanina no caso de Escherichia coli (unha bacteria Gram-negativa) ou L-alanina, D-glutamina, L-lisina, e D-alanina cunha ponte peptídica de 5 glicinas que une os tetrapéptidos no caso de Staphylococcus aureus (unha bacteria Gram-positiva). Estes aminoácidos, excepto os L-aminoácidos, non aparecen en proteínas, polo que poderían ser unha protección contra o ataque da maioría das peptidases.
Os enlaces cruzados entre os aminoácidos de diferentes cadeas de aminoazucres establécense pola acción do encima transpeptidase e orixinan unha estrutura tridimensional forte e ríxida. A secuencia específica de aminoácidos e a estrutura molecular varía coa especie bacteriana considerada. [8]
-
Estrutura do peptidoglicano.
-
Parede celular Gram-positiva.
-
Proteína de unión á penicilina formando enlaces cruzados nunha parede bacteriana acabada de formar.
Inhibición por antibióticos
Some antibacterial drugs such as penicillin interfere with the production of peptidoglycan by binding to bacterial enzymes known as penicillin-binding proteins or transpeptidases.[6] Penicillin-binding proteins form the bonds between oligopeptide crosslinks in peptidoglycan. For a bacterial cell to reproduce through binary fission, more than a million peptidoglycan subunits (NAM-NAG+oligopeptide) must be attached to existing subunits.[9] Mutations in transpeptidases that lead to reduced interactions with an antibiotic are a significant source of emerging antibiotic resistance.[10]
Considered the human body's own antibiotic, lysozymes found in tears work by breaking the β-(1,4)-glycosidic bonds in peptidoglycan (see below) and thereby destroying many bacterial cells. Antibiotics such as penicillin commonly target bacterial cell wall formation (of which peptidoglycan is an important component) because animal cells do not have cell walls.
Biosíntese
The peptidoglycan monomers are synthesized in the cytosol and are then attached to a membrane carrier bactoprenol. Bactoprenol transports peptidoglycan monomers across the cell membrane where they are inserted into the existing peptidoglycan.[11]
In the first step of peptidoglycan synthesis, the glutamine, which is an amino acid, donates an amino group to a sugar, fructose 6-phosphate. This turns fructose 6-phosphate into glucosamine-6-phosphate. In step two, an acetyl group is transferred from acetyl CoA to the amino group on the glucosamine-6-phosphate creating N-acetyl-glucosamine-6-phosphate.[12] In step three of the synthesis process, the N-acetyl-glucosamine-6-phosphate is isomerized, which will change N-acetyl-glucosamine-6-phosphate to N-acetyl-glucosamine-1-phosphate.[12]
In step four, the phosphate-N-acetyl-glucosamine-1-phosphate, which is now a mono phosphate, attacks UTP. Uridine triphosphate, which is a pyrimidine nucleotide, has the ability to act as an energy source. When UDP is used as an energy source, it gives off an inorganic phosphate. In this particular reaction after the monophosphate has attacked the UTP, a phosphate is given off as pyrophosphate, an inorganic phosphate, and is replaced by the monophosphate, creating UDP-Nacetylglucosamine (2,4. This initial stage, is used to create the precursor for the NAG in peptidoglycan.
In step 5, some of the UDP-Nacetylglucosamine (UDP-GlcNAc) is converted to UDP-MurNAc (UDP-N acetylmuramic acid) by the addition of a lactyl group to the glucosamine. Also in this reaction, C3 hydroxyl group will remove a phosphate form the alpha carbon of phosphenol pyruvate. This creates what is called an enol derivative that will be reduced to a “lactyl moiety” by NADPH in step six.[12]
In step 7, the UDP–MurNAc is converted to UDP-MurNAC pentapeptide by the addition of five amino acids, usually including the dipeptide D-alanyl-D-alanine.[12] Each of these reactions requires the energy source ATP.[12] This is all referred to as Stage one.
Stage two is occurs in the cytoplasmic membrane. It is in the membrane where a lipid carrier called bactoprenol carries peptidoglycan precursors through the cell membrane. Bactoprenol will attack the UDP-MurNAc penta, creating a PP-MurNac penta, which is now a lipid. UDP-GlcNAc is then transported to MurNAc, creating Lipid-PP-MurNAc penta-GlcNAc, a disaccharide, also a precursor to peptidoglycan.[12] How this molecule is transported through the membrane is still not understood. However, once it is there, it is added to the growing glycan chain.[12] The next reaction is known as tranglycosylation. In the reaction, the hydroxyl group of the GlcNAc will attach to the MurNAc in the glycan, which will displace the lipid-PP from the glycan chain. The enzyme responsible for this is transglycosylase.[12] Modelo:Clear-left
Notas
- ↑ Animation of Synthesis of Peptidoglycan Layer
- ↑ Madigan, M. T., J. M. Martinko, P. V. Dunlap, and D. P. Clark. Brock biology of microorganisms. 12th ed. San Francisco, CA: Pearson/Benjamin Cummings, 2009.
- ↑ van den Ent F, Amos LA, Löwe J (2001). "Prokaryotic origin of the actin cytoskeleton.". Nature 413 (6851): 39–44. PMID 11544518. doi:10.1038/35092500.
- ↑ van den Ent F, Johnson CM, Persons L, de Boer P, Löwe J (2010). "Bacterial actin MreB assembles in complex with cell shape protein RodZ.". EMBO J 29 (6): 1081–90. PMC 2845281. PMID 20168300. doi:10.1038/emboj.2010.9.
- ↑ C.Michael Hogan. 2010. Bacteria. Encyclopedia of Earth. eds. Sidney Draggan and C.J.Cleveland, National Council for Science and the Environment, Washington DC
- ↑ 6,0 6,1 Salton MRJ, Kim KS (1996). Univ of Texas Medical Branch, ed. Structure. In: Baron's Medical Microbiology (Barron S et al., eds.) (4th ed.). ISBN 0-9631172-1-1.
- ↑ Demchick PH, Koch AL (1 February 1996). "The permeability of the wall fabric of Escherichia coli and Bacillus subtilis". Journal of Bacteriology 178 (3): 768–73. PMC 177723. PMID 8550511.
- ↑ Ryan KJ, Ray CG (editors) (2004). McGraw Hill, ed. Sherris Medical Microbiology (4th ed.). ISBN 0-8385-8529-9.
- ↑ Bauman R (2007). 2nd, ed. Microbiology with Diseases by Taxonomy. Benjamin Cummings. ISBN 0-8053-7679-8.
- ↑ Spratt BG (1994). "Resistance to antibiotics mediated by target alterations". Science (New York) 264 (5157): 388–93. PMID 8153626. doi:10.1126/science.8153626. Parámetro descoñecido
|month=ignorado (Axuda) - ↑ "II. THE PROKARYOTIC CELL: BACTERIA". Consultado o 1. MAY 2011.
- ↑ 12,0 12,1 12,2 12,3 12,4 12,5 12,6 12,7 White, D. (2007). The physiology and biochemistry of prokaryates (3rd ed.). NY: Oxford University Press Inc.